Documente Academic
Documente Profesional
Documente Cultură
Translation
Încărcat de
kvictoTitlu original
Drepturi de autor
Formate disponibile
Partajați acest document
Partajați sau inserați document
Vi se pare util acest document?
Este necorespunzător acest conținut?
Raportați acest documentDrepturi de autor:
Formate disponibile
Translation
Încărcat de
kvictoDrepturi de autor:
Formate disponibile
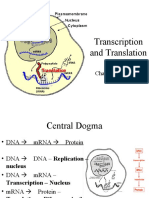
TRANSLATION This is the conversion of the information of mRNA into proteins; messenger RNA produced by transcription is decoded by the
ribosome to produce a specific amino acid chain, or polypeptide, that will later fold into an active protein. Occurs in the ribosomes which exist as dissociate subunit in the cytoplasm and thus have to be assembled together before translation can take place. The ribosome facilitates decoding by inducing the binding of tRNAs with complementary anticodon sequences to that of the mRNA. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through. It involves several steps; Activation In activation, the correct amino acid is covalently bonded to the correct transfer RNA (tRNA). The amino acid is joined by its carboxyl group to the 3' OH of the tRNA by a peptide bond. When the tRNA has an amino acid linked to it, it is termed "charged". Initiation Initiation of translation usually involves the interaction of certain key proteins with a special tag bound to the 5' cap. The protein factors bind the small ribosomal subunit (40S), and these initiation factors hold the mRNA in place. The eukaryotic Initiation Factor 3 (eIF3) is associated with the small ribosomal subunit, and plays a role in keeping the large ribosomal subunit from prematurely binding. eIF3 also interacts with the eIF4F complex which consists of three other initiation factors: eIF4A, eIF4E and eIF4G. eIF4G is a scaffolding protein which directly associates with both eIF3 and the other two components. eIF4E is the cap-binding protein. It is the rate-limiting step of cap-dependent initiation, and is often cleaved from the complex by some viral proteases to limit the cell's ability to translate its own transcripts.
Elongation With the formation of the complex containing fMet-tRNA in the peptidyl site, an aminoacyl tRNA with the complementary anticodon sequence can bind to the mRNA passing through the acceptor site. This binding is aided by elongation factors that are dependent upon the energy from the hydrolysis of GTP. Elongation factors go through a cycle to regenerate GTP after its hydrolysis.
Now, with tRNA bearing a chain of amino acids in the ribosome p site and tRNA containing a single amino acid in the ribosome A site, the addition of a link to the chain can be made. This addition occurs through the formation of a peptide bond, the nitrogen-carbon bond that forms between amino acid subunits to form a polypeptide chain. This bond is catalyzed by the enzyme peptidyl transferase. The peptide bond occurs between the carboxyl group on the lowest link in the peptide chain located at the p site and the amine group on the amino acid in the A group. As a result, the peptide chain shifts over to the A site, with the original amino acid on the A site as the lowest link in the chain. The tRNA in the A site becomes peptidyl RNA, and shifts over to the P site. Meanwhile, the ribosome engages in a process called translocation: spurred by elongation factors, the ribosome moves three nucleotides in the 3' prime direction along the mRNA. In other words, the ribosome moves so that a new mRNA codon is accessible in the A site. Termination Translation ends when one of three stop codons, UAA, UAG, or UGA, enters the A site of the ribosome. There are no aminoacyl tRNA molecules that recognize these sequences. Instead, release factors bind to the P site, catalyzing the release of the completed polypeptide chain and separating the ribosome into its original small and large subunits. POSTTRANSLATIONAL MODIFICATION This is a process which involves chemical modification of a protein after its translation. It is one of the later steps in protein biosynthesis and thus gene expression. During protein synthesis, 20 different amino acids can be incorporated to become a protein. After translation, the posttranslational modification of amino acids extends the range of functions of the protein by attaching it to other biochemical functional groups such as acetate, phosphate, various lipids and carbohydrates, by changing the chemical nature of an amino acid (e.g. citrullination) or by making structural changes, like the formation of disulfide bridges. Also, enzymes may remove amino acids from the amino end of the protein, or cut the peptide chain in the middle. For instance, the peptide hormone insulin is cut twice after disulfide bonds are formed, and a propeptide is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bond.
Figure 2 showing the post translation modification in insulin protein (wikipedia) Differences between translation in Eukaryotes and Prokaryotes RNA is usually translated while it is being transcribed since both transcription and translation takes place in the cytoplasm. Transcription of the eukaryotic mRNA occurs in the nucleus, mRNA then moves to the cytoplasm for translation. Another difference is that eukaryotic mRNA must be modified by capping and intron splicing before it can be used to make protein. However, in prokaryotes, the mRNA usually doesnt need much modification and that is why it is able to undergo translation before transcription is done. Most prokaryotic mRNA molecules often contain transcripts of several genes because of clustering of genes related by functions (operons) thus more than one protein is usually expressed by these mRNas. The Eukaryotic mRNA usually specifies only a single protein.
S-ar putea să vă placă și
- How To Study The Genome GenomeDocument14 paginiHow To Study The Genome GenomekvictoÎncă nu există evaluări
- Structure and Physiology: ProtozoaDocument2 paginiStructure and Physiology: ProtozoakvictoÎncă nu există evaluări
- Assignment MolDocument4 paginiAssignment MolkvictoÎncă nu există evaluări
- 15 ElectrophoresisDocument13 pagini15 ElectrophoresiskvictoÎncă nu există evaluări
- DIscussionDocument6 paginiDIscussionkvictoÎncă nu există evaluări
- Antigenic VariationDocument7 paginiAntigenic VariationkvictoÎncă nu există evaluări
- Ecological ConceptsDocument12 paginiEcological ConceptskvictoÎncă nu există evaluări
- Trypanosomiasis AfricanDocument5 paginiTrypanosomiasis AfricangowindamijayaÎncă nu există evaluări
- MicrobesDocument19 paginiMicrobeskvictoÎncă nu există evaluări
- Gene Expression DianaDocument32 paginiGene Expression DianakvictoÎncă nu există evaluări
- How To Determine Sample SizeDocument5 paginiHow To Determine Sample SizeShaleem DavidÎncă nu există evaluări
- 1269 FullDocument2 pagini1269 FullkvictoÎncă nu există evaluări
- TrypsDocument1 paginăTrypskvictoÎncă nu există evaluări
- Ambrose-Desertation Finalll DraftttttDocument75 paginiAmbrose-Desertation Finalll DraftttttkvictoÎncă nu există evaluări
- Circumsporozoite Protein Genes of Malaria Parasites (Plasmodium SPP.) : Evidence For Positive Selection Immunogenic RegionsDocument9 paginiCircumsporozoite Protein Genes of Malaria Parasites (Plasmodium SPP.) : Evidence For Positive Selection Immunogenic RegionskvictoÎncă nu există evaluări
- Genetic CodeDocument2 paginiGenetic CodekvictoÎncă nu există evaluări
- CBA Brochure IntlDocument16 paginiCBA Brochure IntlkvictoÎncă nu există evaluări
- Preparation of ReagentsDocument4 paginiPreparation of ReagentskvictoÎncă nu există evaluări
- TranscriptionDocument4 paginiTranscriptionkvictoÎncă nu există evaluări
- Preparation of ReagentsDocument4 paginiPreparation of ReagentskvictoÎncă nu există evaluări
- Trypanosome InfectionDocument2 paginiTrypanosome InfectionkvictoÎncă nu există evaluări
- Chapter ThreeDocument2 paginiChapter ThreekvictoÎncă nu există evaluări
- Trypanosome Species and CytokinesDocument1 paginăTrypanosome Species and CytokineskvictoÎncă nu există evaluări
- OmicsDocument6 paginiOmicskvictoÎncă nu există evaluări
- TranscriptionDocument4 paginiTranscriptionkvictoÎncă nu există evaluări
- Homologus RecombinationDocument2 paginiHomologus RecombinationkvictoÎncă nu există evaluări
- African TrypsDocument3 paginiAfrican TrypskvictoÎncă nu există evaluări
- Please Note:: Haemoctymeter Instructions. Haemocytv3.doc DR Neville A. PunchardDocument3 paginiPlease Note:: Haemoctymeter Instructions. Haemocytv3.doc DR Neville A. PunchardkvictoÎncă nu există evaluări
- Gene ExpressionDocument19 paginiGene Expressionkvicto100% (1)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeDe la EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeEvaluare: 4 din 5 stele4/5 (5783)
- The Yellow House: A Memoir (2019 National Book Award Winner)De la EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Evaluare: 4 din 5 stele4/5 (98)
- Never Split the Difference: Negotiating As If Your Life Depended On ItDe la EverandNever Split the Difference: Negotiating As If Your Life Depended On ItEvaluare: 4.5 din 5 stele4.5/5 (838)
- Shoe Dog: A Memoir by the Creator of NikeDe la EverandShoe Dog: A Memoir by the Creator of NikeEvaluare: 4.5 din 5 stele4.5/5 (537)
- The Emperor of All Maladies: A Biography of CancerDe la EverandThe Emperor of All Maladies: A Biography of CancerEvaluare: 4.5 din 5 stele4.5/5 (271)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceDe la EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceEvaluare: 4 din 5 stele4/5 (890)
- The Little Book of Hygge: Danish Secrets to Happy LivingDe la EverandThe Little Book of Hygge: Danish Secrets to Happy LivingEvaluare: 3.5 din 5 stele3.5/5 (399)
- Team of Rivals: The Political Genius of Abraham LincolnDe la EverandTeam of Rivals: The Political Genius of Abraham LincolnEvaluare: 4.5 din 5 stele4.5/5 (234)
- Grit: The Power of Passion and PerseveranceDe la EverandGrit: The Power of Passion and PerseveranceEvaluare: 4 din 5 stele4/5 (587)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaDe la EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaEvaluare: 4.5 din 5 stele4.5/5 (265)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryDe la EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryEvaluare: 3.5 din 5 stele3.5/5 (231)
- On Fire: The (Burning) Case for a Green New DealDe la EverandOn Fire: The (Burning) Case for a Green New DealEvaluare: 4 din 5 stele4/5 (72)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureDe la EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureEvaluare: 4.5 din 5 stele4.5/5 (474)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersDe la EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersEvaluare: 4.5 din 5 stele4.5/5 (344)
- The Unwinding: An Inner History of the New AmericaDe la EverandThe Unwinding: An Inner History of the New AmericaEvaluare: 4 din 5 stele4/5 (45)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyDe la EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyEvaluare: 3.5 din 5 stele3.5/5 (2219)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreDe la EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreEvaluare: 4 din 5 stele4/5 (1090)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)De la EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Evaluare: 4.5 din 5 stele4.5/5 (119)
- Her Body and Other Parties: StoriesDe la EverandHer Body and Other Parties: StoriesEvaluare: 4 din 5 stele4/5 (821)
- Al-Jahiz (781-869) : ZoologyDocument25 paginiAl-Jahiz (781-869) : ZoologyJA QuibzÎncă nu există evaluări
- BMSolutions PDFDocument48 paginiBMSolutions PDF曾hahaÎncă nu există evaluări
- Asianalysis 2009 Kuala Lumpur PosterDocument1 paginăAsianalysis 2009 Kuala Lumpur PosterpeptidesynthesizerÎncă nu există evaluări
- FSC111 - Cell Structure and FunctionDocument15 paginiFSC111 - Cell Structure and FunctionKenneth SilvaÎncă nu există evaluări
- Chapter 22 Nucleic Acids and Protein SynthesisDocument68 paginiChapter 22 Nucleic Acids and Protein SynthesisAa PalmaÎncă nu există evaluări
- Dintzis ExerciseDocument5 paginiDintzis ExerciserichschurÎncă nu există evaluări
- Chemistry Honours RegularDocument72 paginiChemistry Honours RegularSk ArmaanÎncă nu există evaluări
- 14 - Fatty Acid SynthesisDocument5 pagini14 - Fatty Acid SynthesishamedÎncă nu există evaluări
- People To Observe and Describe Microorganisms Generation: LivingDocument7 paginiPeople To Observe and Describe Microorganisms Generation: LivingSabnin SoroniÎncă nu există evaluări
- Lipid BiosynthesisDocument187 paginiLipid BiosynthesisThanh NguyenÎncă nu există evaluări
- Biochemistry Syllabus Amended OnlineDocument3 paginiBiochemistry Syllabus Amended OnlineZach MaxwellÎncă nu există evaluări
- General Bio I - Final Exam - 2013Document7 paginiGeneral Bio I - Final Exam - 2013Yonas Isa'acÎncă nu există evaluări
- Protein synthesis study guide and quizDocument3 paginiProtein synthesis study guide and quizFatimaIjazÎncă nu există evaluări
- Mindoro State College of Agriculture and Technology: I. ObjectivesDocument7 paginiMindoro State College of Agriculture and Technology: I. ObjectivesJunjun CaoliÎncă nu există evaluări
- DNA to protein translationDocument3 paginiDNA to protein translationBernadette Vidon-PangilinanÎncă nu există evaluări
- DNA Replication Fidelity and Strand SynthesisDocument6 paginiDNA Replication Fidelity and Strand SynthesisAlbertoÎncă nu există evaluări
- Pharmacognosy - 2 ThakurDocument290 paginiPharmacognosy - 2 Thakurshoaib1985100% (8)
- SHENIBLOG-Class 10 Biology Focus Area Covered Notes (Eng Med) All Chapters 2022Document16 paginiSHENIBLOG-Class 10 Biology Focus Area Covered Notes (Eng Med) All Chapters 2022kailaskrishnaushusÎncă nu există evaluări
- Carbohydrate Metabolism-1Document27 paginiCarbohydrate Metabolism-1RESHMA I MBAÎncă nu există evaluări
- DNA RNA WorksheetDocument5 paginiDNA RNA WorksheetKathleen FrancoÎncă nu există evaluări
- Ori Hofmekler - Warrior NewsletterDocument49 paginiOri Hofmekler - Warrior NewsletterBigNat7774100% (2)
- Jurnal Fix 1Document16 paginiJurnal Fix 1fitrah fajrianiÎncă nu există evaluări
- Sub: Pharmaceutical Chemistry-II: Topic: SulphonamidesDocument28 paginiSub: Pharmaceutical Chemistry-II: Topic: SulphonamidesRam PrajapatÎncă nu există evaluări
- BIL 255 Exam Questions on DNA-Protein SynthesisDocument31 paginiBIL 255 Exam Questions on DNA-Protein SynthesisMax ArmourÎncă nu există evaluări
- A New Route To Designer AntibioticsDocument2 paginiA New Route To Designer AntibioticsRaphael ContiÎncă nu există evaluări
- Dna Rna Protein Synthesis Homework 1 the Basics of Dna AnswersDocument5 paginiDna Rna Protein Synthesis Homework 1 the Basics of Dna Answersafnaetbpeooztd100% (1)
- AP Bio CH 14 Gene ExpressionDocument24 paginiAP Bio CH 14 Gene ExpressionLuisa RamosÎncă nu există evaluări
- Eukaryotic TranslationDocument24 paginiEukaryotic TranslationCj ScarletÎncă nu există evaluări
- 2nd Peroxisome Metabolism 20760414Document56 pagini2nd Peroxisome Metabolism 20760414Rawbeena RamtelÎncă nu există evaluări
- Plant Cell 1995 Kutchan 1059 70Document13 paginiPlant Cell 1995 Kutchan 1059 70Dawlat SalamaÎncă nu există evaluări