Documente Academic
Documente Profesional
Documente Cultură
Kimball Et Al 2018, Figure 4
Încărcat de
Abby KimballTitlu original
Drepturi de autor
Formate disponibile
Partajați acest document
Partajați sau inserați document
Vi se pare util acest document?
Este necorespunzător acest conținut?
Raportați acest documentDrepturi de autor:
Formate disponibile
Kimball Et Al 2018, Figure 4
Încărcat de
Abby KimballDrepturi de autor:
Formate disponibile
X-shift Settings (VorteX)
A. Dataset Importation:
1. Sample selection:
2. Importation settings:
Select all relevant FCS files
pre-gated for live singlet events
Determine how many events from
each FCS file will be imported
4. Successful dataset importation:
3. Parameters selection: Before clustering the
Select all relevant user should confirm
phenotypic markers that the size and
for clustering dimensions of the
dataset is appropriate 5. Distance Measure:
Select “Angular Distance”
B. Clustering settings:
6. Clustering Algorithm
1. Transformation method: Select the “X-shift algorithm”
Select “arcsinh (x/f)” with f= 5.0
7. Density Estimate
Select the density estimation
2. Noise threshold:
method “N nearest neighbors”
Apply a noise threshold of 1.0
8. Define K values
3. Feature rescaling: Determine what K values will be
Select “none” calculated, (e.g. K values from
5 to 150 at 30 step intervals)
4. Normalization: 9. Number of neighbors:
Select “none” Select “determine automatically”
10. Begin clustering
C. D. Press ‘Go’ to begin clustering
225 Exponential phase calculations (progress of
200
(K=5-K=15) calculations appears in the box)
175
150
Elbow/switch point
# of Clusters
125 (K=20) The elbow/switch point identifies the optimal
number of cell clusters in the dataset, to minimize
100
overfragmentation (exponential phase)
75 Linear phase or underclustering (linear phase).
(K=25-K=150)
Elbow Point Location: 50
The results of the clustering calculations for
25
all given K values can be used to determine an
optimal K value, known as an “elbow point”. 0
150
145
140
135
130
125
120
105
100
115
110
95
90
85
80
75
70
65
60
55
50
45
40
35
30
25
20
15
10
5
0
K value
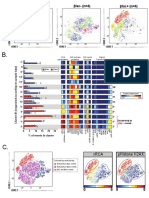
E. The force directed layout generated
Force directed layout settings: in VorteX will look like this and can
1. Events sampled: be colored by cluster ID or expression
5. Edge settings: and can be visualized across individuals
The number of events
The connections or experimental conditions.
from each cluster can
between nodes can
be selected by the user
be determined based
2. Proportional sampling: on node density or by
The events can also be a specific parameter
sampled proportionally
to the cluster size 6. Sample selection:
3. Distance measurement: A force directed layout
can be created from
Choose “angular distance”
one or more samples
4. Num. nearest neighbors:
Use the automatic value
here of 10
F. G. Colored by
phenotype
Colored by
cluster ID
The force directed
layout can be opened
in the Gephi app to
customize figure.
Force directed layout shows
different cell clusters (phenotypes)
spatially distributed based on
CD4+
phenotypic similarity. CD8+
CD19+
CD11b+ CD11c- CD64+
CD11b+ CD11c- CD64-
CD11b+ CD11c+ CD64-
CD45+
CD45-
Clusters can be identified NKp46+
by cluster ID or phenotype.
Figure 4
Kimball et al
S-ar putea să vă placă și
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryDe la EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryEvaluare: 3.5 din 5 stele3.5/5 (231)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)De la EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Evaluare: 4.5 din 5 stele4.5/5 (119)
- Never Split the Difference: Negotiating As If Your Life Depended On ItDe la EverandNever Split the Difference: Negotiating As If Your Life Depended On ItEvaluare: 4.5 din 5 stele4.5/5 (838)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaDe la EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaEvaluare: 4.5 din 5 stele4.5/5 (265)
- The Little Book of Hygge: Danish Secrets to Happy LivingDe la EverandThe Little Book of Hygge: Danish Secrets to Happy LivingEvaluare: 3.5 din 5 stele3.5/5 (399)
- Grit: The Power of Passion and PerseveranceDe la EverandGrit: The Power of Passion and PerseveranceEvaluare: 4 din 5 stele4/5 (587)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyDe la EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyEvaluare: 3.5 din 5 stele3.5/5 (2219)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeDe la EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeEvaluare: 4 din 5 stele4/5 (5794)
- Team of Rivals: The Political Genius of Abraham LincolnDe la EverandTeam of Rivals: The Political Genius of Abraham LincolnEvaluare: 4.5 din 5 stele4.5/5 (234)
- Shoe Dog: A Memoir by the Creator of NikeDe la EverandShoe Dog: A Memoir by the Creator of NikeEvaluare: 4.5 din 5 stele4.5/5 (537)
- The Emperor of All Maladies: A Biography of CancerDe la EverandThe Emperor of All Maladies: A Biography of CancerEvaluare: 4.5 din 5 stele4.5/5 (271)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreDe la EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreEvaluare: 4 din 5 stele4/5 (1090)
- Her Body and Other Parties: StoriesDe la EverandHer Body and Other Parties: StoriesEvaluare: 4 din 5 stele4/5 (821)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersDe la EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersEvaluare: 4.5 din 5 stele4.5/5 (344)
- Spencer - First PrinciplesDocument516 paginiSpencer - First PrinciplesCelephaïs Press / Unspeakable Press (Leng)100% (2)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceDe la EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceEvaluare: 4 din 5 stele4/5 (890)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureDe la EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureEvaluare: 4.5 din 5 stele4.5/5 (474)
- The Unwinding: An Inner History of the New AmericaDe la EverandThe Unwinding: An Inner History of the New AmericaEvaluare: 4 din 5 stele4/5 (45)
- The Yellow House: A Memoir (2019 National Book Award Winner)De la EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Evaluare: 4 din 5 stele4/5 (98)
- Journal Club Presentation PDFDocument2 paginiJournal Club Presentation PDFsiddhartha senÎncă nu există evaluări
- Practical Intuition For SuccessDocument9 paginiPractical Intuition For SuccessMatiÎncă nu există evaluări
- MSA ProcedureDocument3 paginiMSA ProcedureAnkur0% (1)
- Effects of Drug Addiction on the Human BodyDocument19 paginiEffects of Drug Addiction on the Human BodyAshly Galagnara100% (1)
- On Fire: The (Burning) Case for a Green New DealDe la EverandOn Fire: The (Burning) Case for a Green New DealEvaluare: 4 din 5 stele4/5 (73)
- Advantages & Disadvantages of Case StudiesDocument18 paginiAdvantages & Disadvantages of Case StudiesChrestien Pavillar100% (1)
- Jis B 7184 SummaryDocument2 paginiJis B 7184 SummaryVimal100% (1)
- Nursing Research StatisticsDocument7 paginiNursing Research StatisticsAbi RajanÎncă nu există evaluări
- Research DesignDocument11 paginiResearch DesignAastha Vyas0% (1)
- Chapter 3 Morgan Integrating Qualitative and Quantitative Methods 2 PDFDocument18 paginiChapter 3 Morgan Integrating Qualitative and Quantitative Methods 2 PDFcsskwxÎncă nu există evaluări
- Kimball Et Al 2019, Figure 4Document1 paginăKimball Et Al 2019, Figure 4Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 6Document1 paginăKimball Et Al 2019, Figure 6Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 8Document1 paginăKimball Et Al 2019, Figure 8Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 1Document1 paginăKimball Et Al 2019, Figure 1Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 5Document1 paginăKimball Et Al 2019, Figure 5Abby KimballÎncă nu există evaluări
- Tobin Et Al 2019, Figure 2Document1 paginăTobin Et Al 2019, Figure 2Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 2Document1 paginăKimball Et Al 2019, Figure 2Abby KimballÎncă nu există evaluări
- Tobin Et Al 2019, Figure 1Document1 paginăTobin Et Al 2019, Figure 1Abby KimballÎncă nu există evaluări
- Neuwelt Et Al, Figure 2Document1 paginăNeuwelt Et Al, Figure 2Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 7Document1 paginăKimball Et Al 2019, Figure 7Abby KimballÎncă nu există evaluări
- Oko Et Al 2019, Figure 8Document1 paginăOko Et Al 2019, Figure 8Abby KimballÎncă nu există evaluări
- Tobin Et Al, 2019 Figure 3Document1 paginăTobin Et Al, 2019 Figure 3Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 4Document1 paginăKimball Et Al 2019, Figure 4Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 10Document1 paginăKimball Et Al 2018, Figure 10Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 1Document1 paginăKimball Et Al 2019, Figure 1Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 3Document1 paginăKimball Et Al 2019, Figure 3Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 1Document1 paginăKimball Et Al 2018, Figure 1Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 9Document1 paginăKimball Et Al 2018, Figure 9Abby KimballÎncă nu există evaluări
- Bullock Et Al 2018, Supplemental Figure 5Document1 paginăBullock Et Al 2018, Supplemental Figure 5Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 3Document1 paginăKimball Et Al 2018, Figure 3Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 8Document1 paginăKimball Et Al 2018, Figure 8Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 2Document1 paginăKimball Et Al 2018, Figure 2Abby KimballÎncă nu există evaluări
- Bullock Et Al 2018, Figure 6Document1 paginăBullock Et Al 2018, Figure 6Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 7Document1 paginăKimball Et Al 2018, Figure 7Abby KimballÎncă nu există evaluări
- Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectDocument1 paginăGraph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectAbby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 5Document1 paginăKimball Et Al 2018, Figure 5Abby KimballÎncă nu există evaluări
- Berger Et Al 2019, Figure 2Document1 paginăBerger Et Al 2019, Figure 2Abby KimballÎncă nu există evaluări
- Lecture 21: Model Selection 1 Choosing ModelsDocument14 paginiLecture 21: Model Selection 1 Choosing ModelsSÎncă nu există evaluări
- Correction of Chapter-2Document2 paginiCorrection of Chapter-2Hasib IslamÎncă nu există evaluări
- Business Research ProcessDocument15 paginiBusiness Research ProcessDurga Prasad Dash100% (1)
- Conflicts and Solutions in Tempe Lake RevitalizationDocument12 paginiConflicts and Solutions in Tempe Lake RevitalizationFadliadi SadliÎncă nu există evaluări
- Efektivitas Model Pembelajaran Problem Solving Dan Problem Based Learning Ditinjau Dari Kemampuan Pemecahan Masalah Matematika SDDocument14 paginiEfektivitas Model Pembelajaran Problem Solving Dan Problem Based Learning Ditinjau Dari Kemampuan Pemecahan Masalah Matematika SDwebotre netÎncă nu există evaluări
- Practice Exam BHO2285 T2 2018Document4 paginiPractice Exam BHO2285 T2 2018庄敏敏Încă nu există evaluări
- Analyzing M&E Data: Chapter at A GlanceDocument18 paginiAnalyzing M&E Data: Chapter at A GlanceRoqueÎncă nu există evaluări
- Transpersonal Psychology As A Scientific Field: Harris FriedmanDocument13 paginiTranspersonal Psychology As A Scientific Field: Harris Friedman004 Nurul HikmahÎncă nu există evaluări
- Opman Quiz 1 ReviewDocument11 paginiOpman Quiz 1 ReviewReniella AllejeÎncă nu există evaluări
- W9PSDocument9 paginiW9PSpolar necksonÎncă nu există evaluări
- Confronting The Experts - Brian MartinDocument112 paginiConfronting The Experts - Brian Martinragod2100% (1)
- BCS-DS-602: Machine Learning: Dr. Sarika Chaudhary Associate Professor Fet-CseDocument18 paginiBCS-DS-602: Machine Learning: Dr. Sarika Chaudhary Associate Professor Fet-CseAnonmity worldÎncă nu există evaluări
- Imrad FormatDocument5 paginiImrad FormatPeach BubbleÎncă nu există evaluări
- Chapter 4 Statistical Inference in Quality Control and ImprovementDocument106 paginiChapter 4 Statistical Inference in Quality Control and ImprovementEfraim John Hernandez100% (1)
- Test Bank For Statistics For Management and Economics 10th Edition by KellerDocument11 paginiTest Bank For Statistics For Management and Economics 10th Edition by Kellera161855766Încă nu există evaluări
- Practice Set 3 SolutionsDocument2 paginiPractice Set 3 Solutionsbala sanchitÎncă nu există evaluări
- Skittles Project 1Document7 paginiSkittles Project 1api-299868669100% (2)
- A White Paper On Process Improvement in Education: Betty Ziskovsky, MAT Joe Ziskovsky, MBA, CLMDocument19 paginiA White Paper On Process Improvement in Education: Betty Ziskovsky, MAT Joe Ziskovsky, MBA, CLMjoe004Încă nu există evaluări
- Gateways 16e GuidedNotes Chapter1.4Document2 paginiGateways 16e GuidedNotes Chapter1.4Rozita WahabÎncă nu există evaluări
- Stock Price Predictions Using A Geometric Brownian Motion - Joel LidénDocument41 paginiStock Price Predictions Using A Geometric Brownian Motion - Joel LidénMaxÎncă nu există evaluări