Documente Academic
Documente Profesional
Documente Cultură
Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: Correct
Încărcat de
Abby KimballTitlu original
Drepturi de autor
Formate disponibile
Partajați acest document
Partajați sau inserați document
Vi se pare util acest document?
Este necorespunzător acest conținut?
Raportați acest documentDrepturi de autor:
Formate disponibile
Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: Correct
Încărcat de
Abby KimballDrepturi de autor:
Formate disponibile
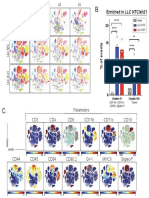
Citrus Settings (Cytobank)
A. 7. Events sampled:
1. Select cellular population: 2. Select markers: 3. Designate experimental groups: 4. Chose association models: Select the number of events
to be sampled from each file
Select cellular population Select all relevant phenotypic Assign FCS files to appropriate Up to three association models can
for analysis markers for clustering experimental groups be used for Citrus analysis (SAM, 8. Cluster size (%):
LASSO/GLMNET, and PAMR) The automatic setting for this is 5%,
but can be changed by the user
5. Cluster characterization:
Clusters can be characterized 9. Cross validation folds:
by cellular abundance or The automatic setting for this is 5,
median expression but can be changed by the user
6. Event sampling: 10. False discovery rate (%):
Events can be sampled The automatic setting for this is 1,
equally or proportionally but can be changed by the user
B. Number of model features Citrus Settings:
0 2 3 4 4 4 6 7 8 9 16 21 25 28 30
Number of characteristics
(populations or markers)
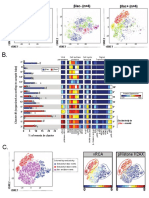
C. Events sampled per file: 9,141
Minimum cluster size: 5%
different between the groups Graph Features: Group Assignment: Random*
100
Cross Validation Error Rate
82218
Number of model features
cv.min model identifies 7 features that Ideally, a model should have low
82251
Feature False Discovery Rate
0 2 2 3 4 7 9 12 13 16 18 20 22 27 35
82246
Model Cross Validation Error Rate
cross validation error rate and
82257
predict differences between groups
100
Cross Validation Error Rate
cv.min
Feature False Discovery Rate 82258
low false discovery rate
82234
cv.min
cv.1se 82247
cv.1se
82238
75
Model Cross Validation Error Rate
82264
75
82248
82265
82262
Citrus defines three predictive cv.fdr.constrained 82256
82267
82252
models with varying stringency
50
82260
CD45
82268 82255 82250
(min, 1se, fdr.constrained) Citrus Settings:
82249
82261
82244 82266 0.93
50
1.72
Events sampled per file: 9,141
25
82263
82245
82259
82253 3.56
82243
82239
Minimum cluster size: 5%
5.02
82254 82236
82240
6.47
0
82227
Group Assignment: Correct
82221 82241
1.12 0.99 0.87 0.74 0.62 0.50 0.37 0.25 0.12 0.00
25
Regularization Threshold
Example of poorly predictive models,
with high cross validation rate and false Note: Despite poor model performance
discovery rate Citrus still identifies clusters
0
4.68 4.16 3.64 3.12 2.60 2.08 1.56 1.04 0.52 0.00
Regularization Threshold Citrus-defined radial hierarchy tree identifies 82238
significant features (from panel B)
D. Citrus Settings: E. Citrus Settings: F. 82258 82253
Events sampled per file: 5,000* Events sampled per file: 9,141 82241
82261
Minimum cluster size: 5% Minimum cluster size: 1%*
Group Assignment: Correct Group Assignment: Correct 82251
82223
Number of model features
82228
82077 82115
0 2 3 4 5 5 8 14 20 31 48 79 114 143
44971
Number of model features 82263
82062 82160
82158
82265
44956 82111 82130
0 1 2 5 5 5 6 7 7 9 15 17 20 24 27
82116
82255
82080 82202 82190
44988
100
82097 82204
44981 Cross Validation Error Rate 82139 82203 82207
82100
Feature False Discovery Rate
82200
44978 82216
100
82195
Cross Validation Error Rate cv.min
82206 82178
82229
82243
82186
82242
Feature False Discovery Rate 44986 44983 cv.1se 82228 82249 82221
82045
82247
cv.min cv.fdr.constrained
82227
82205
82162 82095 82050
82262
Model Cross Validation Error Rate
82163 82257
44993 82168
cv.1se 82267
82229
82240
82148
cv.fdr.constrained
75
82222 82247 82188 81951
82234 82147 82058
Model Cross Validation Error Rate
82262
82246
44984 82201
82259
82117
44997 82255
75
82239
82151
44992 82213
82268 82236 82232
82265 82211
82071 82177
82263
44994 82193 82258 82137
82252
44998 44985 82259 82235
82225 82267
82135 82210
82191
50
82264
82091 82192
44980 82254
82268 82136 82120
82256
82264
44999 44972 44964 82198
82266
82152
82230 82092
44996 82223
CD45
50
82181
81998 82266 82260
81950 82219
82252
81686 82131 82253
44990
82249
82090 82244 82150
CD45
82021
44995 82212
82184
82238
82196
82261
82165 82231
82256 82127
82017
0.41
25
82104 82251
82232
1.56
82164 82248 82217
82215 82067
44989
Model:
82250
44976 82159
44991 82129
82183
82185 82138
1.72 82250
82260
25
44979
2.59
82224 82236
82153 82241 82245
44952 82233
82170
3.36
Min
82101 82208
82220
82141 82189
44987 44982 82180 82199
3.89 82046 82194
82214 82179
82237
82209 82172
4.99 82243
82230
0
82161 82145
82173 82112
82155 82142
SE
82156 82169 82123
44973 44962
5.19
82257
82144 82102
82197
82134 82081
6.62
5.67 5.04 4.41 3.78 3.15 2.52 1.89 1.26 0.63 0.00
82087 82044
0
82149
6.48 82048
Constrained
82254
82176
44968
4.21 3.74 3.27 2.81 2.34 1.87 1.40 0.94 0.47 0.00 Regularization Threshold 81902
82089
Regularization Threshold 82239
Decreasing the minimum Decreasing the minimum cluster 82234
Decreasing the input cell number cluster size to 1% results in size to 1% creates a higher
This Citrus plot looks appropriate, but
results in 10% model error rate 10% model error rate density radial tree There are 7 model features
the model indicates it is not opimal
identfied by the min and SE models There are 25 model features
that predict differences between identfied by the constrained model
experimental groups that predict differences between
Citrus portrays data In a Citrus tree, all events are present in the experimental groups
as a radial tree, but central node, and subjected to further The hierarchical relationship
unlike SPADE all bifurcation. Each cell can be present in between parent and daughters
events are hierarchical. multiple nodes (unlike a SPADE tree). is revealed by subtle arrows.
Colored by Parameter Expression (CD45) Colored by significance models
G. 82238 CD45
2.02
H. 82238
Models have different
levels of strigency Model: Population:
2.97
82258 82253
Min
Background
82258 82253
K.
Parent clusters contain a mixture of cells
82261
4.14
82241
82261
that are subdivided between daughter clusters. SE
Cluster
Tree Geometry
82241
Original Radial
5.32
82223
82251
6.50 82223
82251
Constrained
(Default)
82228
82265 82263
82228
82265 82263 82255
82255
82229
82247
82229
82265
82262
82267
82247 82267
82262
82267
B6 Biased
82259
82259 82268 82236 82232
82268 82236 82232
82264
82256
CD64
82264 82266
CD45
82266 82256 82252
82249
82249 82252
82250
82260
82250
Model:
82260
82230 82243
82257
82230 82243
Min 82257
SE
82263 82251 82264 82265
82254
82254
82243
82239
Constrained 82234
82239
82234
CD45 CD45 CD64 CD64
I. 82268 J. 82268
IL10KO biased
82252 82266
CD4
(Manually generated)
82266 82267 82266 IL10KO biased 82267 B6 Biased
Lag3 CD8
Modifed Vertical
Tree Geometry
82260 82256 82264 82265 82260 82256 82264 82265
82243 82236 82260 82256
CD4
CD4
82230 82257 82252 82250 82259 82263 82251 82230 82257 82252 82250 82259 82263 82251
Lag3 Lag3 CD8 CD8
82239 82254 82243 82236 82249 82255 82262 82261 82241 <5%
* 82239 82254 82243 82236 82249 82255 82262 82261 82241
82234 82232 82228 82247 82258 82234 82232 82228 82247 82258
Citrus trees contain bifurcation Certain markers are discriminators
Model: points in which daughters between divergent daughters.
6.50 5.32 4.14 2.97 2.02 82223 82229 82253 82238
Min 82223 82229 82253 82238 diverge in their significance.
CD45 Expression SE
Constrained
A manually generated The Citrus tree only depicts nodes
vertical tree demonstrates that reach the minimum cluster size
parent-daughter relationships
and clarifies which
(here 5%), daughters with lower Figure 6
frequencies (e.g. in gray) are not shown.
clusters are subgroups. Kimball et al
S-ar putea să vă placă și
- Instant Assessments for Data Tracking, Grade 3: MathDe la EverandInstant Assessments for Data Tracking, Grade 3: MathEvaluare: 5 din 5 stele5/5 (2)
- Panoma (Council Grove) Geomodel Initial Simulations (Single Well)Document1 paginăPanoma (Council Grove) Geomodel Initial Simulations (Single Well)BouregaÎncă nu există evaluări
- Instant Assessments for Data Tracking, Grade 5: MathDe la EverandInstant Assessments for Data Tracking, Grade 5: MathÎncă nu există evaluări
- Differential Privacy of Hierarchical Census Data: An Optimization ApproachDocument17 paginiDifferential Privacy of Hierarchical Census Data: An Optimization ApproachJohn SmithÎncă nu există evaluări
- Gans + Final Practice Questions: Instructor: Preethi JyothiDocument28 paginiGans + Final Practice Questions: Instructor: Preethi JyothiSammy KÎncă nu există evaluări
- Edexcel iGCSE ICT Software MindmapDocument1 paginăEdexcel iGCSE ICT Software MindmapPaul ZambonÎncă nu există evaluări
- Demetra Guideline1Document44 paginiDemetra Guideline1erererefgdfgdfgdfÎncă nu există evaluări
- 840D Ommunication Default SettingDocument5 pagini840D Ommunication Default SettingPatryk MarczewskiÎncă nu există evaluări
- ASIC Implementation of Shared LUT Based Distributed Arithmetic in FIR FilterDocument4 paginiASIC Implementation of Shared LUT Based Distributed Arithmetic in FIR Filtermuneeb rahmanÎncă nu există evaluări
- ScanX Digital Integration Instructions 03.06.2018Document2 paginiScanX Digital Integration Instructions 03.06.2018Rafael CastroÎncă nu există evaluări
- Traffic Assignment Algorithm ComparisonDocument36 paginiTraffic Assignment Algorithm ComparisonSiervo Andrés Aguirre BenavidesÎncă nu există evaluări
- Classified ch1-6Document65 paginiClassified ch1-6snsÎncă nu există evaluări
- Amino Acid Analysis in Plasma-Serum and Urine Brochure en 4Document6 paginiAmino Acid Analysis in Plasma-Serum and Urine Brochure en 4yousrazeidan1979Încă nu există evaluări
- DahmannDocument20 paginiDahmanncoopter zhangÎncă nu există evaluări
- 4 - RESC - Teo - 2122Document49 pagini4 - RESC - Teo - 2122paulo.jcr.oliveiraÎncă nu există evaluări
- Improving Deep Neural Networks: Hyperparameter Tuning, Regularization and OptimizationDocument1 paginăImproving Deep Neural Networks: Hyperparameter Tuning, Regularization and OptimizationSharath Poikayil SatheeshÎncă nu există evaluări
- Distributed Control System: Technical Data SheetDocument13 paginiDistributed Control System: Technical Data Sheetbrome2014Încă nu există evaluări
- Denoising Diffusion Probabilistic ModelsDocument25 paginiDenoising Diffusion Probabilistic Modelsgovas53832Încă nu există evaluări
- Chin Her-7. SLD Inv 1-7Document1 paginăChin Her-7. SLD Inv 1-7uzair rosdiÎncă nu există evaluări
- Xun Huang Multimodal Unsupervised Image-To-Image ECCV 2018 PaperDocument18 paginiXun Huang Multimodal Unsupervised Image-To-Image ECCV 2018 PaperyonasÎncă nu există evaluări
- Quadshot AbstractDocument3 paginiQuadshot AbstractHirak BandyopadhyayÎncă nu există evaluări
- ManagementDocument1 paginăManagementmarcofrfamÎncă nu există evaluări
- STD1 Crimping Chart March 2020 Final 2Document1 paginăSTD1 Crimping Chart March 2020 Final 2Luciano RodriguesÎncă nu există evaluări
- TCP: An Approximation To The Real State Diagram: Nonexist ListenDocument1 paginăTCP: An Approximation To The Real State Diagram: Nonexist Listenatl_avÎncă nu există evaluări
- Font Samples - All SymbolsDocument24 paginiFont Samples - All SymbolsChristianÎncă nu există evaluări
- Elbsound Music Font Samples - All SymbolsDocument26 paginiElbsound Music Font Samples - All Symbolsangerm55Încă nu există evaluări
- FontSamplesAllSymbols PDFDocument26 paginiFontSamplesAllSymbols PDFMattÎncă nu există evaluări
- Rapid Object Detection Using A Boosted Cascade of Simple FeaturesDocument9 paginiRapid Object Detection Using A Boosted Cascade of Simple FeaturesYuvraj NegiÎncă nu există evaluări
- Assignment 4/5 Statistics 5.301 Due: Nov. 29: Rule 1: If One or More of The Next Seven Samples Yield Values of The SampleDocument6 paginiAssignment 4/5 Statistics 5.301 Due: Nov. 29: Rule 1: If One or More of The Next Seven Samples Yield Values of The SamplePatih Agung SÎncă nu există evaluări
- Part III 2019Document72 paginiPart III 2019yilunzhangÎncă nu există evaluări
- Practice Quiz 1 Solutions: Introduction To AlgorithmsDocument9 paginiPractice Quiz 1 Solutions: Introduction To AlgorithmsporkerriaÎncă nu există evaluări
- AyatDocument4 paginiAyatarifin photocopyÎncă nu există evaluări
- 3 - ClusteringDocument39 pagini3 - Clusteringd.pinchukÎncă nu există evaluări
- High-Resolution Radar System Modeling With Matlab/Simulink: ElectronicsDocument7 paginiHigh-Resolution Radar System Modeling With Matlab/Simulink: ElectronicsosamaÎncă nu există evaluări
- Forces and Moments: Beard & Mclain, "Small Unmanned Aircraft," Princeton University Press, 2012 Chapter 4: Slide 1Document43 paginiForces and Moments: Beard & Mclain, "Small Unmanned Aircraft," Princeton University Press, 2012 Chapter 4: Slide 1aslanım kralıçamÎncă nu există evaluări
- CBS 1Document6 paginiCBS 1Rishubh GandhiÎncă nu există evaluări
- 8.1 Worksheet Polar CoordinatesDocument3 pagini8.1 Worksheet Polar CoordinatesRonnieMaeMaullionÎncă nu există evaluări
- DatasheetDocument49 paginiDatasheetRakesh KrÎncă nu există evaluări
- Separators in The Cement IndustryDocument5 paginiSeparators in The Cement Industryzane truesdaleÎncă nu există evaluări
- Road Safety Forecasts in Five European Countries Using Structural Time-Series ModelsDocument1 paginăRoad Safety Forecasts in Five European Countries Using Structural Time-Series ModelsEndashu KeneaÎncă nu există evaluări
- 03 Generative TavenardDocument7 pagini03 Generative Tavenardsafa ameurÎncă nu există evaluări
- Spartan BWDocument1 paginăSpartan BWMOHSENÎncă nu există evaluări
- MAML - Volume 9 - Issue 1 - Pages 385-466Document81 paginiMAML - Volume 9 - Issue 1 - Pages 385-466mamanona012277Încă nu există evaluări
- 10500-indian-Geography-bits-telugu-AJARUDDIN GK GROUPSDocument174 pagini10500-indian-Geography-bits-telugu-AJARUDDIN GK GROUPSanand babuÎncă nu există evaluări
- 04 - Graphs of Trig Functions PDFDocument4 pagini04 - Graphs of Trig Functions PDFMark Abion ValladolidÎncă nu există evaluări
- 04 - 4 - Graphs of Trig FunctionsDocument4 pagini04 - 4 - Graphs of Trig FunctionsSUNGMIN CHOIÎncă nu există evaluări
- Water Mist Layout - PlanDocument1 paginăWater Mist Layout - PlanPrivate PersonÎncă nu există evaluări
- E/R 3Rd-Dk Plan E/R 4Th-Dk Plan Acc. G-DK Acc. Navi.-Dk E/C A-Dk S/Gear Room E/C B-DK E/C C-DKDocument1 paginăE/R 3Rd-Dk Plan E/R 4Th-Dk Plan Acc. G-DK Acc. Navi.-Dk E/C A-Dk S/Gear Room E/C B-DK E/C C-DKMs RawatÎncă nu există evaluări
- Atf Estimation Part1Document41 paginiAtf Estimation Part1mujtabaiftikhar156Încă nu există evaluări
- Proceedings of The 11th Asia Europe WorkDocument45 paginiProceedings of The 11th Asia Europe WorkAndony SanchezÎncă nu există evaluări
- Kerajaan Malaysia: Minconsult Sdn. BHDDocument15 paginiKerajaan Malaysia: Minconsult Sdn. BHDJak CuboÎncă nu există evaluări
- Summer Training ContentDocument3 paginiSummer Training ContentvinayÎncă nu există evaluări
- Rainfall PredictionDocument8 paginiRainfall PredictionANIKET DUBEYÎncă nu există evaluări
- Agile Technologies and SqlalchemyDocument34 paginiAgile Technologies and SqlalchemyColorTwist - Resolve DCTL ToolsÎncă nu există evaluări
- ETM ViewerDocument1 paginăETM Viewerdmitry esaulkovÎncă nu există evaluări
- MGT 3614 - Business - Analytics - Project - Assignment - Phase1: VIT-AP School of BusinessDocument16 paginiMGT 3614 - Business - Analytics - Project - Assignment - Phase1: VIT-AP School of BusinessArjun ArjunÎncă nu există evaluări
- Poster Apr19Document1 paginăPoster Apr19api-312901205Încă nu există evaluări
- 8-Class First Term - First Examination From Biology Lesson ObjectiveDocument1 pagină8-Class First Term - First Examination From Biology Lesson ObjectiveAigerim ImanayevaÎncă nu există evaluări
- So Do Luoi Truyen Tai Dien Viet Nam Nam 2021 (678KB) (678 KB)Document1 paginăSo Do Luoi Truyen Tai Dien Viet Nam Nam 2021 (678KB) (678 KB)taimd2100% (1)
- JR Sanskrit SandhiDocument1 paginăJR Sanskrit SandhipachikollasrinivasaraoÎncă nu există evaluări
- Oko Et Al 2019, Figure 8Document1 paginăOko Et Al 2019, Figure 8Abby KimballÎncă nu există evaluări
- Neuwelt Et Al, Figure 2Document1 paginăNeuwelt Et Al, Figure 2Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 7Document1 paginăKimball Et Al 2019, Figure 7Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 2Document1 paginăKimball Et Al 2019, Figure 2Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 4Document1 paginăKimball Et Al 2019, Figure 4Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 3Document1 paginăKimball Et Al 2019, Figure 3Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 9Document1 paginăKimball Et Al 2018, Figure 9Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 1Document1 paginăKimball Et Al 2019, Figure 1Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 7Document1 paginăKimball Et Al 2018, Figure 7Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 8Document1 paginăKimball Et Al 2018, Figure 8Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 5Document1 paginăKimball Et Al 2018, Figure 5Abby KimballÎncă nu există evaluări
- Bullock Et Al 2018, Figure 6Document1 paginăBullock Et Al 2018, Figure 6Abby KimballÎncă nu există evaluări
- Bullock Et Al 2018, Supplemental Figure 5Document1 paginăBullock Et Al 2018, Supplemental Figure 5Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 1Document1 paginăKimball Et Al 2018, Figure 1Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 2Document1 paginăKimball Et Al 2018, Figure 2Abby KimballÎncă nu există evaluări
- Berger Et Al 2019, Figure 2Document1 paginăBerger Et Al 2019, Figure 2Abby KimballÎncă nu există evaluări
- Wireless Body Area Networks: A SurveyDocument29 paginiWireless Body Area Networks: A Surveyabdul hananÎncă nu există evaluări
- ExcavationDocument18 paginiExcavationBogdan-Gabriel SchiopuÎncă nu există evaluări
- White Products CatalogDocument49 paginiWhite Products CatalogjesusÎncă nu există evaluări
- 2016-10-03 NYCHA Maximo Siebel Status Closed (Audit Trail)Document4 pagini2016-10-03 NYCHA Maximo Siebel Status Closed (Audit Trail)Progress QueensÎncă nu există evaluări
- Building 101 - 25 Tips For A Tropical HomeDocument11 paginiBuilding 101 - 25 Tips For A Tropical HomeMelanie CabforoÎncă nu există evaluări
- Myanmar Power SystemDocument4 paginiMyanmar Power Systemkayden.keitonÎncă nu există evaluări
- 09931374A Clarus 690 User's GuideDocument244 pagini09931374A Clarus 690 User's GuideLuz Idalia Ibarra Rodriguez0% (1)
- A330 Tire Inspn PDFDocument21 paginiA330 Tire Inspn PDFRithesh Ram NambiarÎncă nu există evaluări
- Teacher Evaluation in A Blended Learning EnviornmentDocument11 paginiTeacher Evaluation in A Blended Learning Enviornmentapi-287748301Încă nu există evaluări
- Omni III User's Manual Va.1.0-20140124Document130 paginiOmni III User's Manual Va.1.0-20140124Reuel TacayÎncă nu există evaluări
- KSLCDocument52 paginiKSLCzacklawsÎncă nu există evaluări
- New Process Performance IE4 Motors: Product NotesDocument2 paginiNew Process Performance IE4 Motors: Product NotesCali MelendezÎncă nu există evaluări
- Broch 6700 New 1Document2 paginiBroch 6700 New 1rumboherbalÎncă nu există evaluări
- CatalogDocument42 paginiCatalogOnerom LeuhanÎncă nu există evaluări
- Community Needs Assessments and Sample QuestionaireDocument16 paginiCommunity Needs Assessments and Sample QuestionaireLemuel C. Fernandez100% (2)
- DominosDocument11 paginiDominosApril MartinezÎncă nu există evaluări
- Bank Statement - Feb.2020Document5 paginiBank Statement - Feb.2020TRIVEDI ANILÎncă nu există evaluări
- Infineon IKFW50N65DH5Document15 paginiInfineon IKFW50N65DH5nithinmundackal3623Încă nu există evaluări
- Ict Designation FormDocument3 paginiIct Designation FormAloha Mae ImbagÎncă nu există evaluări
- JKM410 430N 54HL4 (V) F3 enDocument2 paginiJKM410 430N 54HL4 (V) F3 enAmer CajdricÎncă nu există evaluări
- Roxtec CatalogDocument41 paginiRoxtec Catalogvux004Încă nu există evaluări
- Johannes Gensfleisch Zur Laden Zum GutenbergDocument2 paginiJohannes Gensfleisch Zur Laden Zum GutenbergNathan SidabutarÎncă nu există evaluări
- Seismic Data AcquisitionDocument14 paginiSeismic Data Acquisitionjoao kiala100% (1)
- Phase ShifterDocument7 paginiPhase ShifterNumanAbdullahÎncă nu există evaluări
- A330-200 CommunicationsDocument42 paginiA330-200 CommunicationsTarik BenzinebÎncă nu există evaluări
- Human Resources Assistant Resume, HR, Example, Sample, Employment, Work Duties, Cover LetterDocument3 paginiHuman Resources Assistant Resume, HR, Example, Sample, Employment, Work Duties, Cover LetterDavid SabaflyÎncă nu există evaluări
- Times of India - Supply Chain Management of Newspapers and MagazinesDocument18 paginiTimes of India - Supply Chain Management of Newspapers and MagazinesPravakar Kumar33% (3)
- Binthen Motorized Curtain Price List Rev1.0 Jan 2022Document4 paginiBinthen Motorized Curtain Price List Rev1.0 Jan 2022Emil EremiaÎncă nu există evaluări
- Camsco Breaker PDFDocument12 paginiCamsco Breaker PDFMichael MaiquemaÎncă nu există evaluări
- Reynolds EqnDocument27 paginiReynolds EqnSuman KhanalÎncă nu există evaluări
- ChatGPT Money Machine 2024 - The Ultimate Chatbot Cheat Sheet to Go From Clueless Noob to Prompt Prodigy Fast! Complete AI Beginner’s Course to Catch the GPT Gold Rush Before It Leaves You BehindDe la EverandChatGPT Money Machine 2024 - The Ultimate Chatbot Cheat Sheet to Go From Clueless Noob to Prompt Prodigy Fast! Complete AI Beginner’s Course to Catch the GPT Gold Rush Before It Leaves You BehindÎncă nu există evaluări
- Generative AI: The Insights You Need from Harvard Business ReviewDe la EverandGenerative AI: The Insights You Need from Harvard Business ReviewEvaluare: 4.5 din 5 stele4.5/5 (2)
- Scary Smart: The Future of Artificial Intelligence and How You Can Save Our WorldDe la EverandScary Smart: The Future of Artificial Intelligence and How You Can Save Our WorldEvaluare: 4.5 din 5 stele4.5/5 (55)
- ChatGPT Side Hustles 2024 - Unlock the Digital Goldmine and Get AI Working for You Fast with More Than 85 Side Hustle Ideas to Boost Passive Income, Create New Cash Flow, and Get Ahead of the CurveDe la EverandChatGPT Side Hustles 2024 - Unlock the Digital Goldmine and Get AI Working for You Fast with More Than 85 Side Hustle Ideas to Boost Passive Income, Create New Cash Flow, and Get Ahead of the CurveÎncă nu există evaluări
- The Master Algorithm: How the Quest for the Ultimate Learning Machine Will Remake Our WorldDe la EverandThe Master Algorithm: How the Quest for the Ultimate Learning Machine Will Remake Our WorldEvaluare: 4.5 din 5 stele4.5/5 (107)
- 100M Offers Made Easy: Create Your Own Irresistible Offers by Turning ChatGPT into Alex HormoziDe la Everand100M Offers Made Easy: Create Your Own Irresistible Offers by Turning ChatGPT into Alex HormoziÎncă nu există evaluări
- Four Battlegrounds: Power in the Age of Artificial IntelligenceDe la EverandFour Battlegrounds: Power in the Age of Artificial IntelligenceEvaluare: 5 din 5 stele5/5 (5)
- ChatGPT Millionaire 2024 - Bot-Driven Side Hustles, Prompt Engineering Shortcut Secrets, and Automated Income Streams that Print Money While You Sleep. The Ultimate Beginner’s Guide for AI BusinessDe la EverandChatGPT Millionaire 2024 - Bot-Driven Side Hustles, Prompt Engineering Shortcut Secrets, and Automated Income Streams that Print Money While You Sleep. The Ultimate Beginner’s Guide for AI BusinessÎncă nu există evaluări
- Artificial Intelligence & Generative AI for Beginners: The Complete GuideDe la EverandArtificial Intelligence & Generative AI for Beginners: The Complete GuideEvaluare: 5 din 5 stele5/5 (1)
- The AI Advantage: How to Put the Artificial Intelligence Revolution to WorkDe la EverandThe AI Advantage: How to Put the Artificial Intelligence Revolution to WorkEvaluare: 4 din 5 stele4/5 (7)
- Who's Afraid of AI?: Fear and Promise in the Age of Thinking MachinesDe la EverandWho's Afraid of AI?: Fear and Promise in the Age of Thinking MachinesEvaluare: 4.5 din 5 stele4.5/5 (13)
- Artificial Intelligence: The Insights You Need from Harvard Business ReviewDe la EverandArtificial Intelligence: The Insights You Need from Harvard Business ReviewEvaluare: 4.5 din 5 stele4.5/5 (104)
- Your AI Survival Guide: Scraped Knees, Bruised Elbows, and Lessons Learned from Real-World AI DeploymentsDe la EverandYour AI Survival Guide: Scraped Knees, Bruised Elbows, and Lessons Learned from Real-World AI DeploymentsÎncă nu există evaluări
- Working with AI: Real Stories of Human-Machine Collaboration (Management on the Cutting Edge)De la EverandWorking with AI: Real Stories of Human-Machine Collaboration (Management on the Cutting Edge)Evaluare: 5 din 5 stele5/5 (5)
- HBR's 10 Must Reads on AI, Analytics, and the New Machine AgeDe la EverandHBR's 10 Must Reads on AI, Analytics, and the New Machine AgeEvaluare: 4.5 din 5 stele4.5/5 (69)
- Artificial Intelligence: A Guide for Thinking HumansDe la EverandArtificial Intelligence: A Guide for Thinking HumansEvaluare: 4.5 din 5 stele4.5/5 (30)
- T-Minus AI: Humanity's Countdown to Artificial Intelligence and the New Pursuit of Global PowerDe la EverandT-Minus AI: Humanity's Countdown to Artificial Intelligence and the New Pursuit of Global PowerEvaluare: 4 din 5 stele4/5 (4)
- Architects of Intelligence: The truth about AI from the people building itDe la EverandArchitects of Intelligence: The truth about AI from the people building itEvaluare: 4.5 din 5 stele4.5/5 (21)
- AI and Machine Learning for Coders: A Programmer's Guide to Artificial IntelligenceDe la EverandAI and Machine Learning for Coders: A Programmer's Guide to Artificial IntelligenceEvaluare: 4 din 5 stele4/5 (2)
- Quantum Physics: A Beginners Guide to How Quantum Physics Affects Everything around UsDe la EverandQuantum Physics: A Beginners Guide to How Quantum Physics Affects Everything around UsEvaluare: 4.5 din 5 stele4.5/5 (3)
- Creating Online Courses with ChatGPT | A Step-by-Step Guide with Prompt TemplatesDe la EverandCreating Online Courses with ChatGPT | A Step-by-Step Guide with Prompt TemplatesEvaluare: 4 din 5 stele4/5 (4)
- Demystifying Prompt Engineering: AI Prompts at Your Fingertips (A Step-By-Step Guide)De la EverandDemystifying Prompt Engineering: AI Prompts at Your Fingertips (A Step-By-Step Guide)Evaluare: 4 din 5 stele4/5 (1)