Documente Academic
Documente Profesional
Documente Cultură
Kimball Et Al 2018, Figure 9
Încărcat de
Abby KimballTitlu original
Drepturi de autor
Formate disponibile
Partajați acest document
Partajați sau inserați document
Vi se pare util acest document?
Este necorespunzător acest conținut?
Raportați acest documentDrepturi de autor:
Formate disponibile
Kimball Et Al 2018, Figure 9
Încărcat de
Abby KimballDrepturi de autor:
Formate disponibile
A.
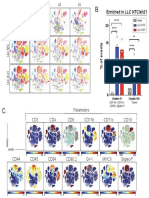
CD11b CD11c Gr-1 PD-L1 Tbet
Gated on: CD4+
Some markers identified visually by
B6 #1
T cells in FlowJo
viSNE are not significantly different.
viSNE
**
IL10KO #1
B.
** 150
Median Intensity
***
ns
*
100
viSNE plots Altered location of CD4+ T cells, according 50
display only to tSNE1 and tSNE2, suggest B6
CD4+ T cell islands IL10KO 0
changes in phenotype of CD4+ T cells.
CD11b CD11c Gr-1 PD-L1 Tbet
C. B6
D. Focused analysis on CD4+ T cell clusters
CD11c CTLA4
Lag3
PhenoGraph defined six
subtypes of CD4+ T cells. PhenoGraph-defined
PhenoGraph
clusters differ in expression
Naive CD4+ T cells, (cluster #16) across multiple parameters.
% CD4+ T cells =16.3% Regulatory CD4+ T cells, (cluster #7)
CD11b ICOS IRF4
Effector CD4+ T cells A, (cluster #12)
IL10KO Effector CD4+ T cells B, (cluster #8)
Effector CD4+ T cells C, (cluster #28)
Effector CD4+ T cells D, (cluster #18) Certain cellular markers
are exclusively positive in
Effector CD4+ T cells A, (cluster #12)
Average of all events
IL10KO mice show IL10KO dominant clusters
Effector CD4+ T cells B, (cluster #8)
increased CD4+ T cells Effector CD4+ T cells C, (cluster #28) (cluster #8 and #28).
and altered distribution
Cluster #524
Individual Events
B6 biased
of CD4+ T cell subtypes. CD4+ T cell clusters that differ in
% CD4+ T cells =30.2%
abundance between conditions.
E. F. Line graphs of a cluster’s median
expression can be used to identify
cell type and phenotype subset.
Cluster #520
Individual Events
8 Cluster #515 Ki6
7 CD1
1b
MH
-1
Median Expression
Sca CI
S
PD1
6
The inner ring indicates core
ICO
IL10KO biased
X-shift
CD3
IRF4
4 CD4+ T cells
markers expressed on all
Tim3
Lag3
#515
1 L
2
44
PD
CD4+ T cells.
CD
Tb CD4 TR
Average of all events e
H.
t GI
Tbet
Tim3
Lag3
IkBa
CD19
NKp46
Ki67
PDL2
CD64
PDL1
IRF4
Sig F
FoxP3
KLRG1
Bcatenin
CD127
CD11c
GITR
MHCI
CD69
CD8
CD11b
CD25
Gr-1
p4eBP1
CD3
CTLA4
PD1
Sca1
CD45
CD44
CD4
CD117
MHCII
ICOS
CTLA4
100 Parameters
8 Cluster #520 IRF
4
-1 MH
Sca
CD
CI
Cellular expression
Median Expression
6
The outer ring defines
MHCII
Gr-1
across all parameters 4
Dendritic cells
X-shift can define the average #520
accessory phenotypes,
showing data from
CD
2
44
11
cellular expression across all
CD
with characteristics
Ki
CD11b
a
c
IkB
67
ten individual cells
parameters (here the most unique to this cell cluster,
Tbet
Tim3
Lag3
IkBa
CD19
NKp46
Ki67
PDL2
CD64
PDL1
IRF4
Sig F
FoxP3
KLRG1
Bcatenin
CD127
CD11c
GITR
MHCI
CD69
CD8
CD11b
CD25
Gr-1
p4eBP1
CD3
CTLA4
PD1
Sca1
CD45
CD44
CD4
CD117
MHCII
ICOS
(in cluster #520) Parameters
IL10KO biased cluster). not consistently present
highlights conserved,
among all cell subsets.
variable phenotypes.
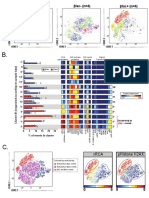
G. CD11b CD11c Gr-1 PD-L1 Tbet H. Node #1, CD4+ T cells
#26
5 ** ns
B6 #1
#1
ns
**
Median Intensity
Changed color on a node 4 ns
indicates altered expression
SPADE
#26 3
IL10KO #1
#1 between conditions.
2
B6
1
1.0 6.5 0.5 6.8 0.3 4.5 0.8 5.8 0.0 4.3
Median expression values IL10KO
for a single node can be 0
Selected CD4+ T cell nodes assessed for significance. CD11b CD11c Gr-1 PDL1 Tbet
K.
Number of model features #82228 IRF4 #82254 IRF4 #82228, PDL1 #82267, CD11b
I. J.
82233 82253
0 2 2 4 4 5 14 26 45 93 206 382 634
**** **** **** ****
Median Expression
Median Expression
Median Expression
Median Expression
82256
3 3 4 6
100
Cross Validation Error Rate Citrus Settings: 82248
3
82239
Feature False Discovery Rate Events sampled per file: 9,141
82261
2 2 4
cv.min
cv.1se
Minimum cluster size: 5%
Group assignment: Correct IRF4 2
PDL1
82265
cv.fdr.constrained
Cluster characterization: Medians 1 1 2
1
82264
Model Cross Validation Error Rate
82228 82249
cv.min model has 10% CD11b
75
82254 82250
0
82258
0
Citrus
0
82267
0
cross validation error rate
6
O
IL 6
O
IL 6
O
IL 6
O
82268 82222
B
B
B
K
82263
K
82260
10
10
10
10
82259
IL
82266
50
82237
82251
82262
82257
#82228
#82254
#82228
#82267
82252
82247
82255
25
82245
82241
IRF4 IRF4 PDL1 CD11b
82217
82220
Population: Background Cluster
Citrus identified 4 features that predict
0
Figure 9
7.15 6.36 5.56 4.77 3.97 3.18 2.38 1.59 0.79 0.00
Regularization Threshold
differences between experimental groups. Kimball et al
S-ar putea să vă placă și
- Solution Manual For Investment Science by David LuenbergerDocument94 paginiSolution Manual For Investment Science by David Luenbergerkoenajax96% (28)
- Gigabyte GA-Z77-DS3H Rev 1.0 BoardView PDFDocument1 paginăGigabyte GA-Z77-DS3H Rev 1.0 BoardView PDFKhánh GiaÎncă nu există evaluări
- Column Shop DrawingDocument1 paginăColumn Shop DrawingdantevariasÎncă nu există evaluări
- Exaudiat Te DominusDocument5 paginiExaudiat Te DominusCatalina ʚïɞÎncă nu există evaluări
- 1 - Light Plot - Overhead - Hairspray - B03Document1 pagină1 - Light Plot - Overhead - Hairspray - B03Mike WoodÎncă nu există evaluări
- Smart Winner New SchematicDocument5 paginiSmart Winner New Schematicscales100% (1)
- XT2129-X - Moto g30 (Capri+) - SB - SchematicsDocument4 paginiXT2129-X - Moto g30 (Capri+) - SB - SchematicsFelipe de san anicetoÎncă nu există evaluări
- Carpark Layout PlanDocument1 paginăCarpark Layout PlanP CyÎncă nu există evaluări
- GA H77 DS3H R1.0 ăÎÍDocument1 paginăGA H77 DS3H R1.0 ăÎÍMinh Nguyen PhuÎncă nu există evaluări
- Dragon Ball Z - Cha-La Head-Cha-LaDocument3 paginiDragon Ball Z - Cha-La Head-Cha-LaJI RAÎncă nu există evaluări
- 740 - Opsb3200-3215-3218Document3 pagini740 - Opsb3200-3215-3218Jessica JacobÎncă nu există evaluări
- v3m BlockedDocument1 paginăv3m BlockedRaúl OjedaÎncă nu există evaluări
- ReleaseNote FileList of X415DA 2009 X64 V1.00Document7 paginiReleaseNote FileList of X415DA 2009 X64 V1.00Karen AdanaqueÎncă nu există evaluări
- 033 - de Colores - C - 02 PDFDocument1 pagină033 - de Colores - C - 02 PDFJan WillemsÎncă nu există evaluări
- PVI 6000 TL OUTD W Quick Installation Guide en RevADocument2 paginiPVI 6000 TL OUTD W Quick Installation Guide en RevA王伯恩Încă nu există evaluări
- A-103 Ground Floor Plan Revised1346586168420Document1 paginăA-103 Ground Floor Plan Revised1346586168420zubair khanÎncă nu există evaluări
- Ta41 40007eDocument2 paginiTa41 40007eJuan CoronelÎncă nu există evaluări
- 51 BEL113 - Longstreet Dixie - Abel - Page - 0033 F Horn 2Document1 pagină51 BEL113 - Longstreet Dixie - Abel - Page - 0033 F Horn 2Giuseppe RufiniÎncă nu există evaluări
- L01 MBLC D05 Fad DWG 07PZ100 ZZ Ele1104 R00Document1 paginăL01 MBLC D05 Fad DWG 07PZ100 ZZ Ele1104 R00Asif SafiÎncă nu există evaluări
- Pmi 635Document3 paginiPmi 635api-3703813Încă nu există evaluări
- El Derecho de Vivir en Paz - Tenor 1Document3 paginiEl Derecho de Vivir en Paz - Tenor 1santiago cerdaÎncă nu există evaluări
- Buseko WarehouseDocument1 paginăBuseko Warehouselenganji simwandaÎncă nu există evaluări
- SMS Nobreak Sen T03395-02 Manager 1400 USBDocument1 paginăSMS Nobreak Sen T03395-02 Manager 1400 USBSamuel LopesÎncă nu există evaluări
- Basisanschlussplan MS4 Sport HPI DDU7 Software Release 36Document1 paginăBasisanschlussplan MS4 Sport HPI DDU7 Software Release 36Sebastian BryceÎncă nu există evaluări
- Footlose: Kenny LogginsDocument2 paginiFootlose: Kenny LogginsMarco PepeÎncă nu există evaluări
- Petronas Carigali SDN BHD: Document Review StatusDocument1 paginăPetronas Carigali SDN BHD: Document Review StatusMohd KhaidirÎncă nu există evaluări
- 87ae5500 Cyr - Uty%Document1 pagină87ae5500 Cyr - Uty%Narendra GadeÎncă nu există evaluări
- SRI JAYAJOTHI UNIT OE STRING LAYOUT 1-ModelDocument1 paginăSRI JAYAJOTHI UNIT OE STRING LAYOUT 1-ModelDIVAKARÎncă nu există evaluări
- Esquema Elétrico Pci Principal: From TransDocument8 paginiEsquema Elétrico Pci Principal: From TransLuiz Clemente PimentaÎncă nu există evaluări
- C-Hmi Server Client: ABB ABBDocument1 paginăC-Hmi Server Client: ABB ABBmahmoud sheblÎncă nu există evaluări
- Fsa 1210 F2Document1 paginăFsa 1210 F2Nagamani ManiÎncă nu există evaluări
- Il Vento D'oro Big Band Arrangement-PianoDocument9 paginiIl Vento D'oro Big Band Arrangement-PianoRobin SveginÎncă nu există evaluări
- 07-22-3971 - USB SchematicDocument1 pagină07-22-3971 - USB SchematicHamiltonÎncă nu există evaluări
- MoliendoDocument1 paginăMoliendofioÎncă nu există evaluări
- Cooling Fand Corolla 2010Document1 paginăCooling Fand Corolla 2010Alfredo jose Medina revattaÎncă nu există evaluări
- Bidp SDH 2022 Ar 119 DtcotDocument1 paginăBidp SDH 2022 Ar 119 DtcotJeffÎncă nu există evaluări
- DWG PDF 20.04.2022Document1 paginăDWG PDF 20.04.2022Sanjan SameerÎncă nu există evaluări
- Ews DrainageDocument1 paginăEws Drainagetahirshaikh143Încă nu există evaluări
- F F+ B GM C: Som: Ritmo: TempoDocument1 paginăF F+ B GM C: Som: Ritmo: TempoAlex OliveiraÎncă nu există evaluări
- Screenshot 2022-10-20 at 3.59.19 PMDocument4 paginiScreenshot 2022-10-20 at 3.59.19 PMphearath.moonengineeringÎncă nu există evaluări
- (Superpartituras - Com.br) EvidenciasDocument1 pagină(Superpartituras - Com.br) EvidenciasEliab Miranda AbÎncă nu există evaluări
- Chitaozinho e Xororo - EvidenciasDocument1 paginăChitaozinho e Xororo - EvidenciasBeatriz CarvalhoÎncă nu există evaluări
- (Superpartituras - Com.br) EvidenciasDocument1 pagină(Superpartituras - Com.br) EvidenciasMarco A. P. AmmirabileÎncă nu există evaluări
- F F+ B GM C: Som: Ritmo: TempoDocument1 paginăF F+ B GM C: Som: Ritmo: TempoBenicio MoriÎncă nu există evaluări
- (Superpartituras - Com.br) EvidenciasDocument1 pagină(Superpartituras - Com.br) EvidenciasCesar SilvaÎncă nu există evaluări
- Chitaozinho e Xororo Evidencias PDFDocument1 paginăChitaozinho e Xororo Evidencias PDFPHERNANDO E KAIKEÎncă nu există evaluări
- Chitaozinho e Xororo Evidencias PDFDocument1 paginăChitaozinho e Xororo Evidencias PDFVinicius MartinsÎncă nu există evaluări
- (Superpartituras - Com.br) EvidenciasDocument1 pagină(Superpartituras - Com.br) EvidenciasPedro Henrique Pezzi BonattoÎncă nu există evaluări
- Legend Barangay Boundary Barangay Road: A C A B C D E FDocument1 paginăLegend Barangay Boundary Barangay Road: A C A B C D E FJoel Rodello B DugeniaÎncă nu există evaluări
- Bidp SDH 2022 Ar 117 DTCMCDocument1 paginăBidp SDH 2022 Ar 117 DTCMCJeffÎncă nu există evaluări
- Est-Casa Club Aires Del Batan-E2Document1 paginăEst-Casa Club Aires Del Batan-E2ISMAEL MONTIELÎncă nu există evaluări
- Suite Violino IDocument2 paginiSuite Violino IDaniel PivaÎncă nu există evaluări
- Oh DarlingDocument3 paginiOh DarlingJoão PedroÎncă nu există evaluări
- Eehc SC MP 0300101Document1 paginăEehc SC MP 0300101ahmedsmman1976Încă nu există evaluări
- As Rev PlanDocument1 paginăAs Rev PlanFadi AlatrashÎncă nu există evaluări
- TCL M66 ChassisDocument10 paginiTCL M66 Chassiskachappilly20120% (1)
- Oneal Opb2020Document5 paginiOneal Opb2020paneletronicaÎncă nu există evaluări
- Sheet Notes: A.5 A.08 C D A.9 B.1N B.9 A.2 B.14 A.1 B.0.5 B.1.9 A.09Document1 paginăSheet Notes: A.5 A.08 C D A.9 B.1N B.9 A.2 B.14 A.1 B.0.5 B.1.9 A.09Cristina Mauren P. PajesÎncă nu există evaluări
- Petronas Carigali SDN BHD: Document Review StatusDocument1 paginăPetronas Carigali SDN BHD: Document Review StatusMohd KhaidirÎncă nu există evaluări
- T460HW03 V5 Bn07-00648a Memc T-ConDocument10 paginiT460HW03 V5 Bn07-00648a Memc T-ConTran Van Thien100% (1)
- Kimball Et Al 2019, Figure 7Document1 paginăKimball Et Al 2019, Figure 7Abby KimballÎncă nu există evaluări
- Oko Et Al 2019, Figure 8Document1 paginăOko Et Al 2019, Figure 8Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 2Document1 paginăKimball Et Al 2019, Figure 2Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 4Document1 paginăKimball Et Al 2019, Figure 4Abby KimballÎncă nu există evaluări
- Neuwelt Et Al, Figure 2Document1 paginăNeuwelt Et Al, Figure 2Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 1Document1 paginăKimball Et Al 2019, Figure 1Abby KimballÎncă nu există evaluări
- Kimball Et Al 2019, Figure 3Document1 paginăKimball Et Al 2019, Figure 3Abby KimballÎncă nu există evaluări
- Bullock Et Al 2018, Supplemental Figure 5Document1 paginăBullock Et Al 2018, Supplemental Figure 5Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 7Document1 paginăKimball Et Al 2018, Figure 7Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 8Document1 paginăKimball Et Al 2018, Figure 8Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 5Document1 paginăKimball Et Al 2018, Figure 5Abby KimballÎncă nu există evaluări
- Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectDocument1 paginăGraph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectAbby KimballÎncă nu există evaluări
- Berger Et Al 2019, Figure 2Document1 paginăBerger Et Al 2019, Figure 2Abby KimballÎncă nu există evaluări
- Bullock Et Al 2018, Figure 6Document1 paginăBullock Et Al 2018, Figure 6Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 2Document1 paginăKimball Et Al 2018, Figure 2Abby KimballÎncă nu există evaluări
- Kimball Et Al 2018, Figure 1Document1 paginăKimball Et Al 2018, Figure 1Abby KimballÎncă nu există evaluări
- Leading The Industry In: Solar Microinverter TechnologyDocument2 paginiLeading The Industry In: Solar Microinverter TechnologydukegaloÎncă nu există evaluări
- EJ Mini Lesson #7Document3 paginiEJ Mini Lesson #7ArmandoÎncă nu există evaluări
- LETRIST Locally Encoded Transform Feature HistograDocument16 paginiLETRIST Locally Encoded Transform Feature HistograHARE KRISHNAÎncă nu există evaluări
- Robot Sensors and TransducersDocument176 paginiRobot Sensors and TransducerssakthivelÎncă nu există evaluări
- NumaticsFRLFlexiblokR072010 EsDocument40 paginiNumaticsFRLFlexiblokR072010 EsGabriel San Martin RifoÎncă nu există evaluări
- Ace Om 230Document3 paginiAce Om 230michaelliu123456Încă nu există evaluări
- 新型重油催化裂化催化剂RCC 1的研究开发Document5 pagini新型重油催化裂化催化剂RCC 1的研究开发Anca DumitruÎncă nu există evaluări
- An Experimental Analysis of Clustering Algorithms in Data Mining Using Weka ToolDocument6 paginiAn Experimental Analysis of Clustering Algorithms in Data Mining Using Weka Toolmishranamit2211Încă nu există evaluări
- Mollier Enthalpy Entropy Chart For Steam - US UnitsDocument1 paginăMollier Enthalpy Entropy Chart For Steam - US Unitslin tongÎncă nu există evaluări
- 2D Pipeline Bottom Roughness - Mochammad ImronDocument6 pagini2D Pipeline Bottom Roughness - Mochammad ImronLK AnhDungÎncă nu există evaluări
- Form 1 ExercisesDocument160 paginiForm 1 Exerciseskays MÎncă nu există evaluări
- ElectrolysisDocument3 paginiElectrolysisRaymond ChanÎncă nu există evaluări
- An FPGA Implementation of A Feed-Back Chaotic Synchronization For Secure CommunicationsDocument5 paginiAn FPGA Implementation of A Feed-Back Chaotic Synchronization For Secure Communicationslaz_chikhi1574Încă nu există evaluări
- All Graphs and Charts Available in Show MeDocument16 paginiAll Graphs and Charts Available in Show MeGANGA TAGRAÎncă nu există evaluări
- ImmunologyDocument8 paginiImmunologyማላያላም ማላያላም89% (9)
- API ISCAN-LITE ScannerDocument4 paginiAPI ISCAN-LITE Scannergrg_greÎncă nu există evaluări
- LTE Rach ProcedureDocument4 paginiLTE Rach ProcedureDeepak JammyÎncă nu există evaluări
- Machine Fault Detection Using Vibration Signal Peak DetectorDocument31 paginiMachine Fault Detection Using Vibration Signal Peak Detectordavison coyÎncă nu există evaluări
- Chapter 1: Coding Decoding: Important Note: It Is Good To Avoid Pre-Defined Coding Rule To Write A Coded MessageDocument7 paginiChapter 1: Coding Decoding: Important Note: It Is Good To Avoid Pre-Defined Coding Rule To Write A Coded MessageUmamÎncă nu există evaluări
- Ce2202 - Mechanics of FluidsDocument3 paginiCe2202 - Mechanics of FluidsPrashant GaradÎncă nu există evaluări
- OcrDocument16 paginiOcrBeena JaiswalÎncă nu există evaluări
- Bruh I Hate File Handling - CPPDocument3 paginiBruh I Hate File Handling - CPPJayson AmodiaÎncă nu există evaluări
- Dbms-Lab Assignment - 1: Name - VIKAS SINGH Roll No - 4257Document3 paginiDbms-Lab Assignment - 1: Name - VIKAS SINGH Roll No - 4257Vikas SinghÎncă nu există evaluări
- Assignment - 1 Introduction of Machines and Mechanisms: TheoryDocument23 paginiAssignment - 1 Introduction of Machines and Mechanisms: TheoryAman AmanÎncă nu există evaluări
- Microgrid Modeling and Grid Interconnection StudiesDocument71 paginiMicrogrid Modeling and Grid Interconnection StudiesVeeravasantharao BattulaÎncă nu există evaluări
- List of Knapsack Problems: 2 Multiple ConstraintsDocument3 paginiList of Knapsack Problems: 2 Multiple ConstraintssaiÎncă nu există evaluări
- Graphite PropertiesDocument42 paginiGraphite PropertiesAnnisa Puspa MustikaÎncă nu există evaluări
- Comparative Study of Steel Structure With and Without Floating Columns in EtabsDocument13 paginiComparative Study of Steel Structure With and Without Floating Columns in EtabsBehroz BehzadÎncă nu există evaluări